Tools
Scientific literature search
CRISPR tools
Tool to predict Bacteriophage defense systems
Tool to predict CRISPR-Cas type and much more
Tool to find CRISPR arrays and Cas genes
Tool to find CRISPR arrays and select spacers
Database of CRISPR arrays
Tool to predict targets of CRISPR spacers
Tool to predict CRISPR arrays and Cas gene systems
Tool to analyze CRISPR repeat structure
Interactive CRISPR suite
Tool for Cas9 and Cpf1 target selection
Genome editing tools (Cas9 / Cas12a)
Various links to genome editing tools
Other tools
IMG - Integrated microbial genome database
STRING - Protein networks and genomic neighbourhood
SMART - Protein domain prediction
Protein calculator - Prediction tool for protein properties (mass etc.)
Phyre 2 – Protein structure prediction
iTasser – Protein structure prediction
iVirus - Virus bioinformatics suite
PDB database - Protein structure
Signal P - Signal peptides in proteins
Weblogo - Motif finding and representation
Blogs and popular Science
Small things considered by Elio Schaechter and Roberto Kolter
Blog about the cool things Microbes do
Virology blog by Vincent Racaniello
Blog about viruses and disease
Scientias in Dutch
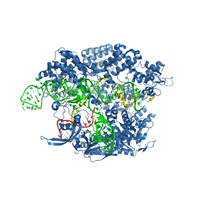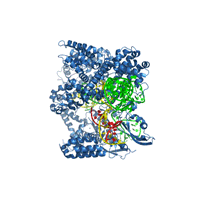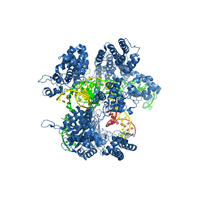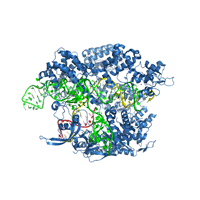
Crystal structure of Cas9 bound to guide RNA and single stranded target DNA
Host institute
Research interest
My lab is interested in the interaction between microbes and bacteriophages. We study the mechanisms that bacteria use to protect themselves from infections including CRISPR and other phage defense systems, and we explore the adaptations that viruses have evolved to avoid defence systems. We isolate and engineer bacteriophages for phage therapy applications.
Funding
Address
Kavli Institute of Nanoscience
Delft University of Technology
Applied Sciences Building (nr. 58) Office E1.480
Van der Maasweg 9
2629 HZ Delft
Netherlands






