Publications
2025
18. Van den Berg D, Costa AR, Esser JQ, Muralidharan A, Van den Bossche H, Brouns SJJ. Discovery of phage defense systems through component modularity networks. BioRxiv 2025.
This preprint describes how you can employ the modularity of phage defense components (for example sensing or effector domains) to discover new phage defense systems. Using this method we uncover Dionysus, Ophion and Ambrosia, the first two of which provide defense against nucleus forming Jumbo phages.
17. Otwinowska A, Olejniczak S, Latka A, Pozniak M, Majkowska-Skrobek G, Maciejewska B, Koszucki J, Panicker V, Jabłońska S, Hulsens M, Bugert JJ, Tournebize R, Brouns SJJ, Squeglia F, Berisio R, Briers Y, Mostowy R and Drulis-Kawa Z. DepoCatalog: Mapping the Diversity of 105 Recombinant Klebsiella Phage Depolymerases Across Sequence, Structure, and Substrate Specificity. BioRxiv 2025.
This preprint presents a catalog of depolymerases used by Klebsiella phages to infect Klebsiella strains with various capsule types.
112. Costa AR, van den Berg DF, Esser JQ, van den Bossche H, Pozhydaieva N, Kalogeropoulos K, Brouns SJJ. Bacteriophage genomes encode both broad and specific counter-defense repertoires to overcome bacterial defense systems. Cell Host Microbe. 2025 Jul 9;33(7):1161-1172.e5.
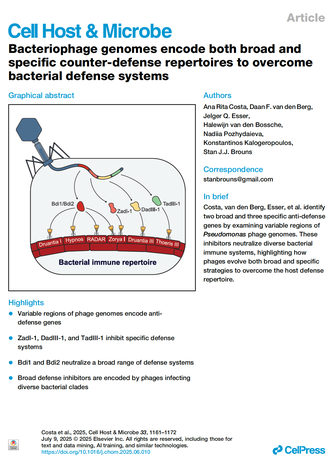
This paper shows the discovery that phage genomes encode multiple inhibitors of phage defense. Some are very specific - inhibiting just one defense system - while others are broad, inhibiting up to four independent defense systems.
16. Muralidharan A, Costa AM, Fierlier D, van den Berg DF, van den Bossche H, Zoumaro-Djayoon AD, Pabst M, Pacesa M, Correia B, Brouns SJJ. Molecular basis for anti-jumbo phage immunity by AVAST Type 5. BioRxiv. 2025
This paper shows how AVAST 5 phage defense systems recognize an early expressed phage protein from Jumbophages to rapidly mount a NAD depletion response.
111. Mahler M, Cui L, Smith LM, Wandera KG, Dietrich O, Mayo-Muñoz D, Balamkundu S, Lee ME, Ye H, Liu CF, Wu J, Mathew J, Dubrulle J, Malone LM, Jackson SA, Fairbanks AJ, Dedon PC, Brouns SJJ, Fineran PC. Phage arabinosyl-hydroxy-cytosine DNA modifications result in distinct evasion and sensitivity responses to phage defense systems. Cell Host Microbe. 2025 Jun 24:S1931-3128(25)00234-3.
110. Van den Berg DF, Brouns SJJ. Phage tRNAs: Decoding the Enigma. Trends in Microbiology.
This work reviews the versatile functions of phage encoded tRNAs, including their role in overcoming phage defense.
109. Van den Berg DF, Brouns SJJ. Reduced prevalence of phage defense systems in Pseudomonas aeruginosa strains from cystic fibrosis patients. mBio. 2025 Feb 25:e0354824.
This work establishes that Cystic Fibrosis derived strains have adapted their phage defense repertoire to their environment.
2024
15. Van den Berg DF, Brouns SJJ. Reduced prevalence of phage defense systems in Pseudomonas aeruginosa strains from cystic fibrosis patients. BioRxiv 2024.
14. Rothschild-Rodriguez D, ..., Nobrega FL. KlebPhaCol: A community-driven resource for Klebsiella research identified a novel gut phage order associated with the human gut. BioRxiv 2024.
108. Van den Berg DF, Costa AR, Esser JQ, Stanciu I, Geissler JQ, Zoumaro-Djayoon AD, Haas PJ, Brouns SJJ. Bacterial homologs of innate eukaryotic antiviral defenses with anti-phage activity highlight shared evolutionary roots of viral defenses. Cell Host Microbe. 2024 Jul 24:S1931-3128(24)00266-X.
This paper identifies over 400 potential phage defense systems predicted from Eukaryotic antiviral systems and validates six new types including Hermes, Prometheus, Erebus, Hypnos, 6A-MBL and Thoeris III.
107. Van Beljouw SPB, Haagsma AC, Kalogeropoulos K, Pabst M, Brouns SJJ. Craspase Orthologs Cleave a Nonconserved Site in Target Protein Csx30. ACS Chem Biol. 2024 May 17;19(5):1051-1055.
This paper reveals the unusual property of the Craspase protease cleaving a non-conserved site of the target protein.
106. Costa AR, van den Berg DF, Esser JQ, Muralidharan A, van den Bossche H, Bonilla BE, van der Steen BA, Haagsma AC, Fluit AC, Nobrega FL, Haas PJ, Brouns SJJ. Accumulation of defense systems in phage-resistant strains of Pseudomonas aeruginosa. Sci Adv. 2024 Feb 23;10(8):eadj0341.
This paper provides the link between phage defense systems and general resistance to phage infection. Strains rich in phage defense systems already circulate among Pseudomonas aeruginosa strains in the clinic
105. van Beljouw SPB, Brouns SJJ. CRISPR-controlled proteases. Biochem Soc Trans. 2024 Feb 28;52(1):441-453.
This paper reviews developments in CRISPR-controlled proteases and focuses on the Craspase family of RNA activated proteases
2023
104. Adler BA, Trinidad MI, Bellieny-Rabelo D, Zhang E, Karp HM, Skopintsev P, Thornton BW, Weissman RF, Yoon PH, Chen L, Hessler T, Eggers AR, Colognori D, Boger R, Doherty EE, Tsuchida CA, Tran RV, Hofman L, Shi H, Wasko KM, Zhou Z, Xia C, Al-Shimary MJ, Patel JR, Thomas VCJX, Pattali R, Kan MJ, Vardapetyan A, Yang A, Lahiri A, Maxwell MF, Murdock AG, Ramit GC, Henderson HR, Calvert RW, Bamert RS, Knott GJ, Lapinaite A, Pausch P, Cofsky JC, Sontheimer EJ, Wiedenheft B, Fineran PC, Brouns SJJ, Sashital DG, Thomas BC, Brown CT, Goltsman DSA, Barrangou R, Siksnys V, Banfield JF, Savage DF, Doudna JA. CasPEDIA Database: a functional classification system for class 2 CRISPR-Cas enzymes. Nucleic Acids Res. 2023 Oct 27:gkad890.
This paper reports CasPEDIA: a curated encyclopedia that integrates enzymatic classification for hundreds of different Cas enzymes across 27 phylogenetic groups spanning the Cas9, Cas12 and Cas13 families, as well as evolutionarily related IscB and TnpB proteins
103. Mahler M, Malone LM, van den Berg DF, Smith LM, Brouns SJJ, Fineran PC. An OmpW-dependent T4-like phage infects Serratia sp. ATCC 39006. Microb Genom. 2023 Mar;9(3):mgen000968.
This paper describes a new T4 like phage for Serratia bacteria
102. van den Berg DF, van der Steen BA, Costa AR, Brouns SJJ. Phage tRNAs evade tRNA-targeting host defenses through anticodon loop mutations. Elife. 2023 Jun 2;12:e85183.
This work presents the hypothesis that phage encoded tRNAs are insensitive to tRNA nuclease activity, a potential antiviral strategy of Bacteria
101. Egido JE, Toner-Bartelds C, Costa AR, Brouns SJJ, Rooijakkers SHM, Bardoel BW, Haas PJ. Monitoring phage-induced lysis of gram-negatives in real time using a fluorescent DNA dye. Sci Rep. 2023 Jan 16;13(1):856.
Paper describing a method to monitor phage infection using fluorescent indicators.
2022
100. Ouyang R, Costa AR, Cassidy CK, Otwinowska A, Williams VCJ, Latka A, Stansfeld PJ, Drulis-Kawa Z, Briers Y, Pelt DM, Brouns SJJ, Briegel A. High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. Nat Commun. 2022 Nov 24;13(1):7241.
This paper shows a 3D reconstruction of a Jumbo phage that uses hyperbranched tail fibers to infect 14 capsular types of Klebsiella pneumoniae
13. Van den Berg DF, Van der Steen BA, Costa AR, Brouns SJJ. Phage tRNAs evade tRNA-targeting host defenses through anticodon loop mutations. 2022 Res sq.
This preprint proposes a new hypothesis: phage tRNAs are resistant to host tRNases
99. van Beljouw SPB, Sanders J, Rodríguez-Molina A, Brouns SJJ. RNA-targeting CRISPR-Cas systems. Nat Rev Microbiol. 2022 Sep 28.
This review provides an overview of the Type III and Type VI CRISPR-Cas systems and the diverse ways how they achieve RNA targeting to stop phage infection.
98. Mahler M, Costa AR, van Beljouw SPB, Fineran PC, Brouns SJJ. Approaches for bacteriophage genome engineering. Trends Biotechnol. 2022 Sep 15.
This work provides an overview of current methods to engineer bacteriophage genomes.
97. Hu C, van Beljouw SPB, Nam KH, Schuler G, Ding F, Cui Y, Rodríguez-Molina A, Haagsma AC, Valk M, Pabst M, Brouns SJJ, Ke A. Craspase is a CRISPR RNA-guided, RNA-activated protease. Science. 2022 Aug 25.
This paper shows how target RNA activates Craspase to switch on protease activity cleaving Csx30
12. Costa AR, Van den Berg DF, Esser JQ, Muralidharan A, Van den Bossche H, Estrada Bonilla B, Van der Steen BA, Haagsma A, Nobrega FL, Haas PJ, Brouns SJJ. Accumulation of defense systems in phage resistant strains of Pseudomonas aeruginosa. BioRxiv 2023.
This work shows the correlation between strains with high numbers of phage defense systems in their genome and high degrees of phage resistance
96. Swartjes T, Shang P, van den Berg DTM, Künne T, Geijsen N, Brouns SJJ, van der Oost J, Staals RHJ, Notebaart RA. Modulating CRISPR-Cas Genome Editing Using Guide-Complementary DNA Oligonucleotides. CRISPR J. 2022 Jul 18.
This paper shows how antisense DNA oligo's can be used to modulate the activity of genome editing nucleases
95. Wu WY, Jackson SA, Almendros C, Haagsma AC, Yilmaz S, Gort G, van der Oost J, Brouns SJJ, Staals RHJ. Adaptation by Type V-A and V-B CRISPR-Cas Systems Demonstrates Conserved Protospacer Selection Mechanisms Between Diverse CRISPR-Cas Types. CRISPR J. 2022 Jul 12.
This work gives insight into the requirements of spacer acquisition and PAM selection in Type V CRISPR-Cas systems, in particular Cas4.
94. Bravo JPK, Aparicio-Maldonado C, Nobrega FL, Brouns SJJ, Taylor DW. Structural basis for broad anti-phage immunity by DISARM. Nat Commun. 2022 May 27;13(1):2987.
This work gives gives structural insights into how the DISARM antiviral complex recognizes free end of viral DNA using a unique trigger loop.
11. Swartjes T, Shang P, Van den Berg D, Künne TA, Geijsen N, Brouns SJJ, Van der Oost J, Staals RHJ, Notebaart RA. Modulating CRISPR-Cas genome editing using guide-complementary DNA oligonucleotides. BioRxiv
93. McKenzie RE, Keizer EM, Vink JNA, van Lopik J, Büke F, Kalkman V, Fleck C, Tans SJ, Brouns SJJ. Single cell variability of CRISPR-Cas interference and adaptation. Mol Syst Biol. 2022 Apr;18(4):e10680.
This paper shows that the timing of primed CRISPR adaptation highly variable: fast in some cells and slow in others
92. Koopal B, Potocnik A, Mutte SK, Aparicio-Maldonado C, Lindhoud S, Vervoort JJM, Brouns SJJ, Swarts DC. Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA. Cell. 2022 Mar 29:S0092-8674(22)00315-4.
This paper shows how a new an RNA guided defense system called SPARTA can deplete NAD(P) cofactors in the cell to trigger suicide
10. Ouyang R, Costa AR, Cassidy CK, Otwinowska A, Williams VCJ, Latka A, Stansfeld PJ, Drulis-Kawa Z, Briers Y, Pelt DM, Brouns SJJ, Briegel A. High resolution reconstruction of a Jumbo bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. BioRxiv
91. Egido JE, Costa AR, Aparicio-Maldonado C, Haas PJ, Brouns SJJ. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol Rev. 2022 Feb 9;46(1):fuab048.
This paper reviews the arsenal of bacteriophage defense systems and its relevance for bacteriophage therapy.
9. Bravo J.P.K, Aparicio-Maldonado C, Nobrega FL, Brouns SJJ, Taylor D. Structural basis of broad phage immunity of DISARM. 2022. BioRxiv
Preprint showing the intricate structure and function of the DrmA and DrmB core complex and its clever way to load invader DNA and perhaps prevent autoimmunity
8. Aparicio-Maldonado C, Ofir G, Salini A, Sorek R, Nobrega FL, Brouns SJJ. Class I DISARM provides anti-phage and anti-conjugation activity by unmethylated DNA recognition. 2022. BioRxiv
This preprint describes how Class I DISARM in Serratia bacteria protect against phage infection and plasmid conjugation, and identifies the role of two cognate sequence motifs in enhancing the recognition of invading DNA
2021
90. Vink JNA, Baijens JHL, Brouns SJJ. PAM-repeat associations and spacer selection preferences in single and co-occurring CRISPR-Cas systems. Genome Biol. Sept 30.
Paper finding more that 30% matches with spacers in the metagenome databases, 200 PAMs, evidence for spacer sharing between CRISPR-Cas types and PAM prediction based on the repeats.
89. Hu C, Almendros C, Nam KH, Costa AR, Vink JNA, Haagsma AC, Bagde SR, Brouns SJJ, Ke A. Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas. Nature. 2021 Sep 29.
Work explaining how PAM selection and directional integration is achieved by Cas4-proteins during CRISPR adaptation
88. van Beljouw SPB, Haagsma AC, Rodríguez-Molina A, van den Berg DF, Vink JNA, Brouns SJJ. The gRAMP CRISPR-Cas effector is an RNA endonuclease complexed with a caspase-like peptidase. Science. 2021 Aug 26:eabk2718.
Breakthrough paper describing the all fused gRAMP and Craspase in Type III-E systems
87. Estrada Bonilla B, Costa AR, van den Berg DF, van Rossum T, Hagedoorn S, Walinga H, Xiao M, Song W, Haas PJ, Nobrega FL, Brouns SJJ. Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae. DNA Res. 2021 Aug 25;28(4):dsab013.
This paper describes four previously unidentified phages from our collection against Klebsiella pneumoniae pathogens. Remarkably the genomes of these phages barely match anything. Kp24, a jumbo phage, is now the founder of the genus VanLeeuwenhoek viridae, and family of Flower viruses, after their branched tail fiber structures
86. Steens JA, Zhu Y, Taylor DW, Bravo JPK, Prinsen SHP, Schoen CD, Keijser BJF, Ossendrijver, Hofstra LM, Brouns SJJ, Shinkai A, van der Oost J, Staals RHJ. SCOPE enables type III CRISPR-Cas diagnostics using flexible targeting and stringent CARF ribonuclease activation. Nat Commun 12, 5033 (2021).
Paper describing how Type III CRISPR-Cas systems can be used for molecular diagnostics of COVID-19
85. Kieper SN, Almendros C, Haagsma AC, Barendregt A, Heck AJR, Brouns SJJ. Cas4-Cas1 Is a Protospacer Adjacent Motif-Processing Factor Mediating Half-Site Spacer Integration During CRISPR Adaptation. CRISPR J. 2021 Aug;4(4):536-548.
This work shows that Cas4 proteins from a complex with Cas1 and capture and process PAM sequences in 3' overhangs to be integrated as new spacers in CRISPR arrays.
7. McKenzie RE, Keizer EM, Vink JNA, van Lopik J, Büke F, Kalkman V, Fleck C, Tans SJ, Brouns SJJ. Single Cell Variability of CRISPR-Cas Interference and Adaptation. 2021. BioRxiv
This paper describes how variable CRISPR-Cas response can be, perhaps resulting in only subpopulations surviving phage infection. The others may be too slow.
6. Vink JNA, Baijens HNA, Brouns SJJ. Comprehensive PAM prediction for CRISPR-Cas systems reveals evidence for spacer sharing, preferred strand targeting and conserved links with CRISPR repeats. BioRxiv.
This paper describes how large scale spacer mapping against metagenomic databases reveals a catalog of PAM, curates CRISPR array orientation, finds evidence for spacer sharing and preferred strand targeting.
84. Castelein SM, Aarts TF, Schleppi J, Hendrikx R, Böttger AJ, Benz D, Marechal M, Makaya A, Brouns SJJ, Schwentenwein M, Meyer AS, Lehner BAE. Iron can be microbially extracted from Lunar and Martian regolith simulants and 3D printed into tough structural materials. PLoS One. 2021 Apr 28;16(4):e0249962.
83. Spoelstra WK, Jacques JM, Gonzalez-Linares R, Nobrega FL, Haagsma AC, Dogterom M, Meijer DH, Idema T, Brouns SJJ, Reese L. CRISPR-based DNA and RNA detection with liquid-liquid phase separation. Biophys J. 2021 Apr 6;120(7):1198-1209. doi: 10.1016/j.bpj.2021.02.013. Epub 2021 Feb 20. PMID: 33617832; PMCID: PMC8059199.
This work shows that liquid phase separation can be used for molecular diagnostics purposes.
5. Estrada Bonilla B, Costa AR, van Rossum T, Hagedoorn S, Walinga H, Xiao M, Song W, Haas P, Nobrega FL, Brouns SJJ. Genomic characterization of four novel bacteriophages infecting the clinical pathogen Klebsiella pneumoniae. BioRxiv.
Paper describing the isolation and genome sequence of four new bacteriophages from sewage water treatment plants. This will expand our collection of bacteriophages against clinical pathogens.
2020
82. Nobrega FL, Walinga H, Dutilh BE, Brouns SJJ. Prophages are associated with extensive CRISPR-Cas auto-immunity. Nucleic Acids Res. 2020 Nov 21:gkaa1071. doi: 10.1093/nar/gkaa1071.
This work shows the extent of self targeting by CRISPR systems, and its link to prophages in the genome of the host.
81. Soliman MYM, Medema G, Bonilla BE, Brouns SJJ, van Halem D. Inactivation of RNA and DNA viruses in water by copper and silver ions and their synergistic effect. Water Res X. 2020 Nov 5;9:100077.
80. de Jonge PA, Smit Sibinga DJC, Boright OA, Costa AR, Nobrega FL, Brouns SJJ, Dutilh BE. Development of styrene maleic acid lipid particles (SMALPs) as a tool for studies of phage-host interactions. J Virol. 2020 Sep 16:JVI.01559-20. doi: 10.1128/JVI.01559-20. Epub ahead of print. PMID: 32938760.
This paper highlights the utility of styrene nanodisks to capture and study bacteriophage receptors in the membrane.
79. Vink JNA, Brouns SJJ, Hohlbein J. Extracting Transition Rates in Particle Tracking Using Analytical Diffusion Distribution Analysis. Biophys J. 2020 Nov 17;119(10):1970-1983.
This paper describes a new data analysis concept to extract protein motion information from live cell microscopy data.
78. de Jonge PA, von Meijenfeldt FAB, Costa AR, Nobrega FL, Brouns SJJ, Dultilh BE. Adsorption Sequencing as a Rapid Method to Link Environmental Bacteriophages to Hosts. iScience. 2020 Aug 6;23(9):101439.
AdsorpSeq will identify novel bacteriophages on the basis of adsorption to their host cell.
4. Vink JNA, Brouns SJJ, Hohlbein J 2020 Extracting transition rates in single-particle tracking using analytical diffusion distribution analysis. BioRxiv. Accepted in Biophys J.
This preprint describes a new concept to analyse data from live cell particle tracking experiments.
3. Nobrega FL, Wallinga H, Dutilh B, Brouns SJJ 2020 Prophages are associated with extensive CRISPR-Cas autoimmunity. BioRxiv. Accepted in NAR.
This preprint contains an overview of self-targeting spacers in all CRISPR-Cas systems and proposes a scenario that self-targeting spacers are acquired against temperate bacteriophages in the prophage state.
77. Kim S, Loeff L, Colombo S, Jergic S, Brouns SJJ, Joo C. Selective loading and processingof prespacers for precise CRISPR adaptation. Nature. 2020. Feb 19.
This paper solves the mystery how spacers are selected based on the PAM, processed by host factors, and integrated in the correct orientation to give functional CRISPR memory and interference.
76. Salazar AN, Nobrega FL, Anyansi C, Aparicio-Maldonado C, Costa AR, Haagsma AC,
Hiralal A, Mahfouz A, McKenzie RE, van Rossum T, Brouns SJJ, Abeel T. An
educational guide for nanopore sequencing in the classroom. PLoS Comput Biol.
2020 Jan 23;16(1):e1007314.
This paper is an educational guide to Nanopore sequencing and bioinformatics for undergraduate students
75. Vink JNA, Martens KJA, Vlot M, McKenzie RE, Almendros C, Estrada Bonilla B,
Brocken DJW, Hohlbein J, Brouns SJJ. Direct Visualization of Native CRISPR Target
Search in Live Bacteria Reveals Cascade DNA Surveillance Mechanism. Mol Cell.
2020 Jan 2;77(1):39-50.e10.
This milestone work shows how Cascade complexes find targets in the bacterial cell.
2019
74. Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2019 Dec 19. doi: 10.1038/s41579-019-0299-x.
This review presents the latest classification of CRISPR-Cas systems
73. Jonge PA, Meijenfeldt FABV, Rooijen LEV, Brouns SJJ, Dutilh BE. Evolution of BACON Domain Tandem Repeats in crAssphage and Novel Gut Bacteriophage Lineages. Viruses. 2019 Nov 21;11(12).
This work analyses the evolution of BACON domains in crAssphage and hosts
72. Cameron P, Coons MM, Klompe SE, Lied AM, Smith SC, Vidal B, Donohoue PD, Rotstein T, Kohrs BW, Nyer DB, Kennedy R, Banh LM, Williams C, Toh MS, Irby MJ, Edwards LS, Lin CH, Owen ALG, Künne T, van der Oost J, Brouns SJJ, Slorach EM, Fuller CK, Gradia S, Kanner SB, May AP, Sternberg SH. Harnessing type I CRISPR-Cas systems for genome engineering in human cells. Nat Biotechnol. 2019 Nov 18.
This paper shows how you can engineer Cascade complexes to be genome editing nucleases in mammalian cells.
71. Martens KJA, van Beljouw SPB, van der Els S, Vink JNA, Baas S, Vogelaar GA,
Brouns SJJ, van Baarlen P, Kleerebezem M, Hohlbein J. Visualisation of dCas9
target search in vivo using an open-microscopy framework. Nat Commun. 2019 Aug
7;10(1):3552.
This work shows how Cas9 surveils all DNA in the bacterial cell to find the target.
70. Lehner BAE, Janssen VAEC, Spiesz EM, Benz D, Brouns SJJ, Meyer AS, van der
Zant HSJ. Creation of Conductive Graphene Materials by Bacterial Reduction Using
Shewanella oneidensis. ChemistryOpen. 2019 Jul 4;8(7):888-895.
This paper describes how you can use microbes to create conductive graphene materials.
69. Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, Cinek O, Aziz
RK, McNair K, Barr JJ, Bibby K, Brouns SJJ, Cazares A, de Jonge PA, Desnues C,
Díaz Muñoz SL, Fineran PC, Kurilshikov A, Lavigne R, Mazankova K, McCarthy DT,
Nobrega FL, Reyes Muñoz A, Tapia G, Trefault N, Tyakht AV, Vinuesa P, Wagemans J,
Zhernakova A, Aarestrup FM, Ahmadov G, Alassaf A, Anton J, Asangba A, Billings
EK, Cantu VA, Carlton JM, Cazares D, Cho GS, Condeff T, Cortés P, Cranfield M,
Cuevas DA, De la Iglesia R, Decewicz P, Doane MP, Dominy NJ, Dziewit L, Elwasila
BM, Eren AM, Franz C, Fu J, Garcia-Aljaro C, Ghedin E, Gulino KM, Haggerty JM,
Head SR, Hendriksen RS, Hill C, Hyöty H, Ilina EN, Irwin MT, Jeffries TC, Jofre
J, Junge RE, Kelley ST, Khan Mirzaei M, Kowalewski M, Kumaresan D, Leigh SR,
Lipson D, Lisitsyna ES, Llagostera M, Maritz JM, Marr LC, McCann A,
Molshanski-Mor S, Monteiro S, Moreira-Grez B, Morris M, Mugisha L, Muniesa M,
Neve H, Nguyen NP, Nigro OD, Nilsson AS, O'Connell T, Odeh R, Oliver A, Piuri M,
Prussin Ii AJ, Qimron U, Quan ZX, Rainetova P, Ramírez-Rojas A, Raya R, Reasor K,
Rice GAO, Rossi A, Santos R, Shimashita J, Stachler EN, Stene LC, Strain R,
Stumpf R, Torres PJ, Twaddle A, Ugochi Ibekwe M, Villagra N, Wandro S, White B,
Whiteley A, Whiteson KL, Wijmenga C, Zambrano MM, Zschach H, Dutilh BE. Global
phylogeography and ancient evolution of the widespread human gut virus
crAssphage. Nat Microbiol. 2019 Jul 8.
This community study analyzed the presence of crAssphage in the gut of individual all over the world and found it to be present in 50% of the global population.
68. Kieper SN, Almendros C, Brouns SJJ. Conserved motifs in the CRISPR leader
sequence control spacer acquisition levels in Type I-D CRISPR-Cas systems. FEMS
Microbiol Lett. 2019 Jun 1;366(11).
This work identifies conserved sequence motifs in the leader sequence that are important for spacer acquisition in Type I-D CRISPR systems.
67. Almendros C, Nobrega FL, McKenzie RE, Brouns SJJ. Cas4-Cas1 fusions drive
efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res. 2019
Apr 2.
This paper shows how efficient Cas4-Cas1 fusion proteins are in selecting PAM compatible spacers in a new type I-U CRISPR-Cas system
66. Musharova O, Sitnik V, Vlot M, Savitskaya E, Datsenko KA, Krivoy A, Fedorov I,
Semenova E, Brouns SJJ, Severinov K. Systematic analysis of Type I-E Escherichia
coli CRISPR-Cas PAM sequences ability to promote interference and primed
adaptation. Mol Microbiol. 2019 Mar 15.
This paper shows the importance of the PAM choice for direct interference and priming.
65. van Sluijs L, van Houte S, van der Oost J, Brouns SJJ, Buckling A, Westra ER.
Addiction systems antagonize bacterial adaptive immunity. FEMS Microbiol Lett.
2019 Mar 5.
This paper investigates the effect of toxin production (CcdB) by F-type conjugative plasmids on CRISPR interference.
64. Almendros C, Kieper SN, Brouns SJJ. CRISPR-Cas systems reduced to a minimum. Mol Cell. 2019 Feb 21. Invited commentary on mini Cas1 and compact Cas9 papers.
63. McKenzie RE, Almendros C, Vink JNA, Brouns SJJ. Using CAPTURE to detect spacer
acquisition in native CRISPR arrays. Nat Protoc. 2019 Feb 11.
This paper details PCR protocols to detect CRISPR adaptation with a very high sensitivity, meaning that you can detect it even when it is happening in a small subpopulation of cells.
2018
2. Martens KJA, Van Beljouw S, van der Els S, Baas S, Vink JNA, Brouns SJJ, Van Baarlen P, Kleerebezem M, Hohlbein J. An open microscopy framework suited for tracking dCas9 in live bacteria. 2018 Oct 8.
1. Spoelstra WK, Jacques JM, Nobrega FL, Haagsma AC, Dogterom M, Idema T, Brouns SJJ, Reese L. CRISPR-based DNA and RNA detection with liquid phase separation. BioRxiv 2018 Nov 20.
This paper describes a novel concept to read out collateral activity of CRISPR nucleases to detect specific nucleic acid sequences.
62. De Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE. Molecular and Evolutionary
Determinants of Bacteriophage Host Range. Trends Microbiol. 2018 Sep 1.
This review discusses the factors determining phage host range
61. Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Dutilh
BE, Brouns SJJ. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol.
2018 Aug 13.
Comprehensive review of phage receptor interactions.
60. Künne T, Zhu Y, da Silva F, Konstantinides N, McKenzie RE, Jackson RN, Brouns
SJJ. Role of nucleotide identity in effective CRISPR target escape mutations.
Nucleic Acids Res. 2018 Aug 11.
This publication shows that the type of mismatch (C or G) in the CRISPR target site strongly affects Cascade binding, and therefore interference and priming efficiency.
59. Costa AR, Brouns SJJ, Nobrega FL. Complete Genome Sequences of Two T4-Like
Escherichia coli Bacteriophages. Genome Announc. 2018 Jun 28;6(26).
58. Loeff L, Brouns SJJ, Joo C. Repetitive DNA Reeling by the Cascade-Cas3 Complex in Nucleotide Unwinding Steps. Mol Cell. 2018 Apr 26.
This publication beautifully shows at the single molecule level how Cas3 is tightly bound to Cascade while reeling in and degrading target DNA
57. Kieper SN, Almendros C, Behler J, McKenzie RE, Nobrega FL, Haagsma AC, Vink
JNA, Hess WR, Brouns SJJ. Cas4 Facilitates PAM-Compatible Spacer Selection during
CRISPR Adaptation. Cell Rep. 2018 Mar 27;22(13):3377-3384.
This breakthrough paper reveals the role of Cas4 in selecting functional instead of random spacers during CRISPR adaptation
56. Vlot M, Houkes J, Lochs SJA, Swarts DC, Zheng P, Kunne T, Mohanraju P, Anders
C, Jinek M, van der Oost J, Dickman MJ, Brouns SJJ. Bacteriophage DNA
glucosylation impairs target DNA binding by type I and II but not by type V
CRISPR-Cas effector complexes. Nucleic Acids Res. 2018 Jan 25;46(2):873-885.
This paper describes how covalent modifications of bacteriophage DNA protects phages from type I and II CRISPR-Cas systems.
55. Vlot M, Nobrega FL, Wong CFA, Liu Y, Brouns SJJ. Complete Genome Sequence of
the Escherichia coli Phage Ayreon. Genome Announc. 2018 Jan 11;6(2). pii:
e01354-17.
2017
54. Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ.
CRISPR-Cas: Adapting to change. Science. 2017 Apr 7;356(6333). Epub 2017 Apr 6.
This review paper summarises our understanding of how microbes change their CRISPR memories of invading viruses and plasmids.
53. Staals RH, Jackson SA, Biswas A, Brouns SJJ, Brown CM, Fineran PC.
Interference-driven spacer acquisition is dominant over naive and primed
adaptation in a native CRISPR-Cas system. Nat Commun. 2016 Oct 3;7:12853.
This paper describes that target interference is coupled to CRISPR adaptation in Type I-F CRISPR-Cas systems in Pectobacterium atrosepticum.

52. Künne T, Kieper SN, Bannenberg JW, Vogel AI, Miellet WR, Klein M, Depken M, Suarez-Diez M, Brouns SJJ. Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation. Mol Cell. 2016 Aug 18.
This paper describes how CRISPR systems cleverly couple target interference to CRISPR memory update. The Cas3 nuclease fragments invader DNA into pieces of near-spacer length enriched for PAM sequences in their 3' ends to form ideal spacer precursors.
51. Xue C, Seetharam AS, Musharova O, Severinov K, J Brouns SJJ, Severin AJ, Sashital DG. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015 Dec 15;43(22):10831-47.
This paper describes how direct interference and priming are affected by spacer choice. In addition, the paper describes how CRISPR arrays avoid self-priming.
50. van der Oost J, Brouns SJJ. CRISPR sabotage. Genome Biol. 2015 Nov 9;16(1):248.
49. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015 Nov;13(11):722-36.
48. Künne T, Westra ER, Brouns SJJ. Electrophoretic Mobility Shift Assay of DNA and CRISPR-Cas Ribonucleoprotein Complexes. Methods Mol Biol. 2015; 1311:171-84.
47. Blosser TR, Loeff L, Westra ER, Vlot M, Künne T, Sobota M, Dekker C, Brouns SJJ, Joo C. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell. 2015 Apr 2;58(1):60-70.
This paper describes how Cascade recognizes targets using two distinct binding modes (PAM-seed dependent and PAM-seed independent) to trigger either direct interference or priming.
46. Swarts DC, Westra ER, Brouns SJJ, van der Oost. Purification and sequencing of DNA guides from prokaryotic Argonaute. Bio-protocol. 2014. 4(22).
45. Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014 Sep 19;345(6203):1473-9.
This paper presents the crystal structure of the Cascade complex from E. coli.
44. Fineran PC, Gerritzen MJ, Suárez-Diez M, Künne T, Boekhorst J, van Hijum SAFT, Staals RHJ, Brouns SJJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):E1629-38.
This paper reveals how direct interference and priming processes in deal with mutations in the target site.
43. Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders APL, Wang Y, Patel DJ, Berenguer J, Brouns SJJ, Van der Oost J. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014 Mar 13;507(7491):258-61
This paper describes DNA-dependent DNA targeting by bacterial Argonaute proteins.
42. Künne T, Swarts DC, Brouns SJJ. Planting the seed: target recognition of short guide RNAs. Trends Microbiol. 2014 Feb;22(2):74-83.
This review highlights the importance of seed sequences in the initial interaction of small RNAs with their targets.
41. Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJJ. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9(9):e1003742.
This paper shows that PAM recognition by Cascade does not take place by direct interactions of the crRNA 5' handle with the PAM in target DNA.
40. Quax TEF, Wolf YI, Koehorst JJ, Wurtzel O, Van der Oost R, Ran W, Blombach F, Makarova KS, Brouns SJJ, Forster AC, Wagner EGH, Sorek R, Koonin EV, Van der Oost J. Differential translation tunes uneven production of operon-encoded proteins. Cell Rep. 2013 Sep 12;4(5):938-44.
This paper reveals how the translation of operon encoded proteins, for example Cascade, is regulated by codon usage.
39. Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lähteinen T, Brouns SJJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013;9(8):e1003683.
38. Ruigrok VJB, Westra ER, Brouns SJJ, Escudé C, Smidt H, Van der Oost J. A capture approach for supercoiled plasmid DNA using a triplex-forming oligonucleotide. Nucleic Acids Res. 2013 May 1;41(10):e111.
This paper presents a method to capture double stranded DNA by forming a triple helix.
37. Westra ER, Staals RHJ, Gort G, Høgh S, Neumann S, de la Cruz F, Fineran PC, Brouns SJJ. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biol. 2013 May;10(5):749-61.
This paper describes the observation that the E. coli CRISPR system is able to target conjugative plasmids.
36. Biswas A, Gagnon JN, Brouns SJJ, Fineran PC, Brown CM. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013 May;10(5):817-27.
This paper describes a great tool for CRISPR target identification and PAM prediction.
35. Westra ER, Swarts DC, Staals RHJ, Jore MM, Brouns SJJ, Van der Oost J. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311-39. Review
34. Westra ER, Nilges B, van Erp PB, Van der Oost J, Dame RT, Brouns SJJ. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012 Sep;9(9):1134-8.
33. Van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJJ, Doudna JA, Van der Oost J, Heck AJR. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics. 2012 Nov;11(11):1430-41.
32. Brouns SJJ. Molecular biology. A Swiss army knife of immunity. Science. 2012. Aug 17;337(6096):808-9. Commentary
This is the accompanying commentary about the seminal paper by Charpentier and Doudna describing the function and programmability of the Cas9, which has opened the door for genome editing applications.
31. Westra ER, Brouns SJJ. The rise and fall of CRISPRs: dynamics of spacer acquisition and loss. Mol Microbiol. 2012 Sep;85(6):1021-5.
30. Swarts DC, Mosterd C, van Passel MW, Brouns SJJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7(4):e35888.
This paper describes naive and primed spacer acquisition in E. coli.
29. Westra ER, van Erp PB, Künne T, Wong SP, Staals RHJ, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, Van der Oost J. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012 Jun 8;46(5):595-605.
This paper shows that Cas3 is recruited to target-bound Cascade complexes to degrade the DNA.
28. Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, Van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011 Sep 21;477(7365):486-9.
27. Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, Van der Oost J, Brouns SJJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):10098-103.
This paper reveals the existence and importance of seed sequences in crRNA.
26. Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, Van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011 Jun;9(6):467-77.
25. Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, Van der Oost J, Brouns SJJ. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011 May;18(5):529-36.
This paper shows how crRNA effector complexes recognize target DNA by forming R-loops. Furthermore, this paper gives some first insights into the structural topology and stoichiometry of the Cascade complex from E. coli.
24. Jore MM, Brouns SJJ, Van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol. 2012 Jun 1;4(6). pii: a003657. Review.
23. Al-Attar S, Westra ER, Van der Oost J, Brouns SJJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011 Apr;392(4):277-89. Review
22. Blombach F, Brouns SJJ, Van der Oost J. Assembling the archaeal ribosome: roles for translation-factor-related GTPases. Biochem Soc Trans. 2011 Jan;39(1):45-50.
21. de Koning B, Blombach F, Brouns SJJ, Van der Oost J. Fidelity in archaeal information processing. Archaea. 2010 Sep 5;2010. pii: 960298. Review
20. Westra ER, Pul Ü, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EGH, Schnetz K, Van der Oost J, Wagner R, Brouns SJJ. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010 Sep;77(6):1380-93.
Commentary by: Mojica FJM, Díez-Villaseñor C. The on–off switch of CRISPR immunity against phages in Escherichia coli, Mol Microbiol. 2010, 77, 6
19. Van der Oost J, Brouns SJJ. RNAi: prokaryotes get in on the act. Cell. 2009 Nov 25;139(5):863-5.
18. Wu H, Sun L, Blombach F, Brouns SJJ, Snijders APL, Lorenzen K, van den Heuvel RHH, Heck AJ, Fu S, Li X, Zhang XC, Rao Z, Van der Oost J. Structure of the ribosome associating GTPase HflX. Proteins. 2010 Feb 15;78(3):705-13.
17. Van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009 Aug;34(8):401-7. Review
16. de Koning B, Blombach F, Wu H, Brouns SJJ, Van der Oost J. Role of multiprotein bridging factor 1 in archaea: bridging the domains? Biochem Soc Trans. 2009 Feb;37(Pt1):52-7. Review
15. Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, Van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008 Aug 15;321(5891):960-4.
This paper describes the discovery of the Cascade complex, how crRNAs are formed by Cas6 and loaded into the Cascade complex. It also shows that Cas3 is required for phage resistance. The mechanism is proposed that CRISPR systems act on the DNA level.
Perspective in Science, Secret Weapon, Young RF. 2008. 321, 922-3
Molecular Biology Select in Cell, CRISPR insights into prokaryotic antiviral defense, Bao MZ. 2008. 135, 7
Research Highlight in Nat Struct Mol Biol, Cascade defense, Chen I, Eggleston AK, Finkelstein JM, Lall S. 2008. 15, 901
Faculty of 1000 Biology
14. Machielsen R, Leferink NG, Hendriks A, Brouns SJJ, Hennemann HG, Daussmann T, Van der Oost J. Laboratory evolution of Pyrococcus furiosus alcohol dehydrogenase to improve the production of (2S,5S)-hexanediol at moderate temperatures. Extremophiles. 2008 Jul;12(4):587-94.
13. Brouns SJJ, Barends TR, Worm P, Akerboom J, Turnbull AP, Salmon L, Van der Oost J. Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily. J Mol Biol. 2008 May 30;379(2):357-71.
12. Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJJ, van de Werken HJG,
Bothner B, Douglas T, van de Oost J, Young MJ. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008 May;82(10):4874-83.
11. Brouns SJJ, Turnbull AP, Willemen HLDM, Akerboom J, Van der Oost J. Crystal structure and biochemical properties of the D-arabinose dehydrogenase from Sulfolobus solfataricus. J Mol Biol. 2007 Aug 31;371(5):1249-60.
10. Wu H, Sun L, Brouns SJJ, Fu S, Akerboom J, Li X, Van der Oost J. Purification, crystallization and preliminary crystallographic analysis of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Mar 1;63(Pt 3):239-41.
9. Van den Heuvel RHH, van Duijn E, Mazon HFM, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, Van der Oost J, Hoyes J, Heck AJR. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal Chem. 2006 Nov 1;78(21):7473-83.
8. Brouns SJJ, Walther J, Snijders APL, van de Werken HJG, Willemen HL, Worm P, de Vos MGJ, Andersson A, Lundgren M, Mazon HFM, van den Heuvel RHH, Nilsson P, Salmon L, de Vos WM, Wright PC, Bernander R, Van der Oost J. Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J Biol Chem. 2006 Sep 15;281(37):27378-88.
This paper reveals a pentose sugar degradation pathway in hyperthermophilic archaea.
Faculty of 1000 Biology
7. Berrisford JM, Hounslow AM, Akerboom J, Hagen WR, Brouns SJJ, Van der Oost J, Murray IA, Michael Blackburn G, Waltho JP, Rice DW, Baker PJ. Evidence supporting a cis-enediol-based mechanism for Pyrococcus furiosus phosphoglucose isomerase. J Mol Biol. 2006 May 19;358(5):1353-66.
6. Brouns SJJ, Smits N, Wu H, Snijders APL, Wright PC, de Vos WM, Van der Oost J. Identification of a novel alpha-galactosidase from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2006 Apr;188(7):2392-9.
5. Snijders APL, Walther J, Peter S, Kinnman I, de Vos MGJ, van de Werken HJG, Brouns SJJ, Van der Oost J, Wright PC. Reconstruction of central carbon metabolism in Sulfolobus solfataricus using a two-dimensional gel electrophoresis map, stable isotope labelling and DNA microarray analysis. Proteomics. 2006 Mar;6(5):1518-29.
4. Gödde C, Sahm K, Brouns SJJ, Kluskens LD, Van der Oost J, de Vos WM, Antranikian G. Cloning and expression of islandisin, a new thermostable subtilisin from Fervidobacterium islandicum, in Escherichia coli. Appl Environ Microbiol. 2005 Jul;71(7):3951-8.
3. Brouns SJJ, Wu H, Akerboom J, Turnbull AP, de Vos WM, Van der Oost J. Engineering a selectable marker for hyperthermophiles. J Biol Chem. 2005 Mar 25;280(12):11422-31.
This paper presents a directed evolution and in vivo selection approach to create a thermostable antibiotic resistance marker for (hyper) thermophilic bacteria and archaea.
Faculty of 1000 Biology
2. Berrisford JM, Akerboom J, Brouns SJJ, Sedelnikova SE, Turnbull AP, Van der Oost J, Salmon L, Hardré R, Murray IA, Blackburn GM, Rice DW, Baker PJ. The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis. J Mol Biol. 2004 Oct 22;343(3):649-57.
1. Kaper T, Brouns SJJ, Geerling AC, De Vos WM, Van der Oost J. DNA family shuffling of hyperthermostable beta-glycosidases. Biochem J. 2002 Dec 1;368(Pt2):461-70.
Publications in Books
7. Nobrega FL, Lavigne R, Brouns SJJ. Revival of Phage therapy. In: Mighty Microbes. the amazing world of microorganisms. Eds. Oudega B et al. Oct 23, 2017.
This illustrated book aims to show that microorganisms are essential for our health, environment, the Earth and its sustainable future. Our chapter describes the past, present and future of bacteriophage therapy.
6. Künne T, Westra ER, Brouns SJJ. Electrophoretic mobility shift assay of DNA and CRISPR-Cas ribonucleoprotein complexes. In Lundgren M, Fineran P, Charpentier E (ed.) In: Methods in Molecular Biology. Springer Verlag, 2015.
5. Staals RHJ, Brouns SJJ. Distribution and mechanism of Type I CRISPR-Cas systems. In: Barrangou R, Van der Oost J (ed.) CRISPR-Cas Immune systems, Springer Verlag, 2013. 145-69. Review
4. Westra ER, Jore MM, Al-Attar S, Brouns SJJ, Van der Oost J. Small RNAs in Bacteria. Encyclopedia of Biological Chemistry 2nd Edition. Elsevier. 2013. 249-55. Review
Westra ER, Van der Oost J, Brouns SJJ (2011) McGraw-Hill Encyclopedia of Science and Technology 2010, 10th ed. 63-6. Review
3. Jore MM, Brouns SJJ, Van der Oost J RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. In: Atkins JF, Gesteland RF and Cech TR (ed.), RNA Worlds: from life's origins to diversity in gene regulation. Cold Spring Harbor Laboratory Press. 2010. 231-42. Review
2. Van de Werken HJGG, Brouns SJJ, and Van der Oost J. Pentose metabolism in Archaea. In: Blum P (ed.) Archaea: new models for prokaryotic biology. Horizon Scientific Press. 2008. 71-94. Review
1. Brouns SJJ, Ettema TJG, Stedman KM, Walther J, Smidt H, Snijders APL, Young M, Bernander R, Wright PC, Siebers B, Van der Oost J. The hyperthermophilic archaeon Sulfolobus solfataricus: from exploration to exploitation. In: Inskeep WP and McDermott TR (ed.), Geothermal biology and geochemistry in Yellowstone national park. Thermal Biology Institute, Montana State University, Bozeman, United States. 2005. 261-76. Review
DISCLAIMER
ANY PAPERS WILL IMMEDIATELY BE REMOVED FROM DOWNLOAD UPON REQUEST
Host institute
Research interest
My lab is interested in the interaction between microbes and bacteriophages. We study the mechanisms that bacteria use to protect themselves from infections including CRISPR and other phage defense systems, and we explore the adaptations that viruses have evolved to avoid defence systems. We isolate and engineer bacteriophages for phage therapy applications.
Funding
Address
Kavli Institute of Nanoscience
Delft University of Technology
Applied Sciences Building (nr. 58) Office E1.480
Van der Maasweg 9
2629 HZ Delft
Netherlands






